Cladistic Taxonomy
Origin of cladistics dates back to early 1900AD but it was Willi Henning in 1960s who coined the term cladistics/phylogenetic systematics, he also developed methodology related to cladistic analysis.
Cladistics is a biological classification system which groups organisms on the basis of shared ancestral characters. It takes into account the evolutionary relatedness e.g. all the organisms having hair are placed in one group called as mammals. It helps in establishing groups of species with the help of similarities and difference in morphological, anatomical, genetic and molecular traits of species. It is used to trace the ancestry and evolution of various traits.
A group of species which share the ancestral characters is called a clade. A clade is strictly a monophylectic group. It can be small or large which depends on the ancestral characters taken into consideration, e.g. all vertebrates are included in one clade based on the presence if vertebral column and when the character taken into consideration is feathers all vertebrates except birds are excluded from the clade. Vertebrates form a larger clade whereas birds form a smaller clade.
The diagramatic representation of the evolutionary relationship among the species is called as Cladogram . In other words Cladograms are the diagrams which are used to demonstrate the evolutionary relationship between the species based on similarities and differences in their physical and genetic traits. Cladograms are also referred to as Phylogenetic trees or Evolutionary tree. But there does exist considerable distinction between them as phylogenetic tree also takes into account the time period taken for evolutionary steps which is represented by long or short distance, distance also depicts the degree of relatedness i.e. short distance means the two or more species are closely related and long distance depicts distant relation between concerned species where as the cladogram simply depicts the evolutionary relationships among organisms.
All life on the earth is a part of single phylogenetic tree. Phylogenetic tree can be of two types Rooted and unrooted. In rooted type we know the ancestral species but in the unrooted tree we don't have the known ancestral species. Rooted phylogenetic trees are always better forms where as the unrooted trees are vague and less informative.
Fig. Different parts of Cladogram
Cladogram consistis of the root which represents the common ancestral species of all Taxa. Divergence of the species from the root due to the evolutionary process leads to the branches which may be internal or external depending upon their positions. Nodes are points on the cladogram which represent common ancestry. A, B, C, D and E represent the terminal points.
Cladograms are constructed by taking into consideration a particular trait. Cladograms constructed by taking into account the morphological characters may differ from the ones which are constructed by taking into consideration the genetic traits. Nowadays advanced computational methods help us take multiple traits into consideration simultaneously for constructing more accurate cladograms i.e.
Cladograms have become more precise and accurate due the advancement in various techniques such as PCR and computational modelling.
They are very useful in depecting the evolutionary history of the lineage of the species, in addition, it is also useful in determining the degree of relatedness between the species .
Cladogram are of two types viz Monophylectic Cladogram and Paraphyletic Cladogram. Monophyletic Cladogram takes into account the ancestral species and all of its descendant species where as paraphyletic cladogram takes into account the ancestral species and some of its descendant species i.e. it doesn't take into account all of the descendent species.
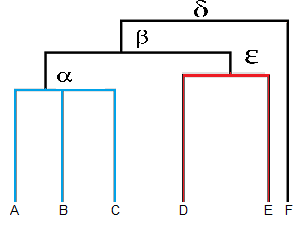
In a Cladogram the clades which are at same level share more traits than those which are at different levels. In the below mentioned Cladogram Delta , beta , alpha and epsilon represent four different clades. A, B, C, D, E and F are the living species which represent the tips.
Fig Cladogram
Species A, B and C share more similarities to each other than they share to species D, E or F. Similarly species F is quite different from species A, B, C, D and E and species D and E are more closely related to each other than they are to A, B, C or F.
In cladistics the traits of any species are categorised as derived and ancestral.
Derived traits are those which have been acquired through the evolutionary process and are not found in the remote ancestral species and ancestral traits are those traits which were also present in the remote ancestral species i.e. they are such traits which have passed on from the ancestral species to the descendent species.
In cladistics we come across many terms whose knowledge is of utmost importance for any taxonomist. They have been described as under:
Apomorphy: It refers to a derived or a specialised trait that is found in a group and is unique to it. Such characters have been found to be evolved in the recent ancestors. These characters are not seen in the distant/remote ancestors e.g. feathers in birds, Hair in mammals, vertebral column of vertebrates and pantadactyle in vertebrates are apomorphic characters . These characters appeared during evolutionary process and are not found in remote ancestors.
Synapomorphy : It is the phenomenon sharing a derived trait by two or more decendent species and their recent ancestal species. It is an apomorphy that is shared by two or more species or groups. Synapomorphy forms the basis of cladistic classification. It helps in establishing the the monophylectic groups or clades.
Underlying synapomarphy : It is character which is a synapomorphy for a clade but for lineages in this clade is A plesiomorphy that is altered in some lineages .
Plesiomorphy: these are the ancestral traits found in various species. Infact these traits have got evolved in distant ancestors and passed on through many evolutionary steps to various descendents e.g. vertebrae in case of man cow and dog is a plesiomorphy as they have evolved in very distant ancestors which was a fish like organisms. In contrast apomorphy represents the derived trait which is evolved in recent ancestors. Plesiomorphy does not help in establishing clade and hence is not helpful in cladistics classification.
Smyplesiomorphy : It is the phenomenon of sharing an ancestral trait by two or more modern groups. In other words it included the characters shared by two or more groups and their earliest or remotest common ancestor. It does not help in establishing the evolutionary relationship between the recent ancestors.
An apomorphy for a larger clade can be a plesiomorphy for a smaller nested clade with in that larger clade e.g. vertebrates form a larger clade where as the mammals form a smaller clade which is a part of vertebrate clade , here the mammalian clade is also called as nested clade, if we talk of vertebral column in vertebrate clade it is a apomorphic character but if we talk of same trait in mammalian clade it is a plesiomorphic character.
Pseudoplesiomorphy : such a character which can't be identified as plesiomorphy or apomorphy.
Homoplasy: it is a character or trait which is found in two distantly related groups, such characters are not found in their common ancestors . Homoplasic characters evolve independently due to similar environmental conditions and thus represent the convergent evolution. e.g. eyes in different animals have evolved independently are an excellent example of Homoplasy. The warmbloodedness in aves and mammals is another example of Homoplasy as this trait has evolved independently in both groups and is not found in their common ancestors.
Autapomorphy: It is a phenomenon of having a derived character in only one species i.e. It is a case of apomorphy restricted to a single species. Speech is an example of autapomorphy as it is restricted to only one species i.e. human beings. Bipedal movement in man in relation ro other primates is another example of autapomorphy but bipedalism in relation to birds is an example of Homoplasy. Autapomorphy does not provide any information about the phylogeny.
Phylogram is a phylogenetic tree whose branch lengths reflect the extant of changes, characters have undergone during the evolution i.e. smaller the length of branch lesser is the deviation from the ancestral species and larger the length of branch large is the deviation and from the ancestral species.
____________ depicts less changes over the period of time
____________________depicts more changes over the period of time .
Chronogram is a phylogenetic tree whose branch lengths are proportional to the time.
Phenetic classification involves grouping of organisms on the basis of observed physical similarities where as Phylogenetic classification involves grouping of organisms based on evolutionary decent.
Phenogram is a branched tree diagramatic representation of phenetic classification which portrays the taxonomic relationship between living organisms of two or more groups. Phenogram does not focus on evolutionary relationship like Cladogram. It lays emphasis on similarities of traits among living Texa without regard to phylogenetic relationship.
Regards
Dr Rahul Kait
Assistant Professor
Department of Zoology
GGM SC COLLEGE JAMMU





Nice going.....it's going to be very Beneficial for the students especially Preparing for all competitive exams....All the best for all ur future endeavours
ReplyDeleteThanks a lot for appreciation
Delete